scikit-learn consortium : performance & parallel computing

May 28 of this year, the scikit-learn consortium took place at Rueil Malmaison
This day was the occasion for me to discover the new features and trends of the Python community when speaking of Machine Learning. As I was convinced I could learn a lot from this day, I showed up with a blank notebook, ready to grab any useful information from the talks.
Disclaimer
This post’s main objective is to gather my notes, as well as my conclusions on the future of performant Machine Learning in Python. Its pretention is certainly not to express the exact way of thinking of the speakers, but rather to provide a subjective interpretation of what has emerged from this day, starting from my notes.
ONNX – Xavier Duprès (Microsoft)
Xavier Duprès goes first for a talk on ONNX (Open Neural Network eXchange), which, as its name implies, was initially designed to improve inter-operability between the existing Deep Learning frameworks.
Some time as passed since then, and the original ONNX project shifted from a pure Deep Learning technology to a more open eco-system, now accepting « classic » ML models. We will thus keep a focus on the sub-part of ONNX that is dedicated to scikit-learn, which provides a high-level API for simplified import/export of pre-trained models
But first, a bit of history …
The joys of pickling
Back in the days when ONNX did not exist (not that long ago you’ll admit), the Python community already had its own serialization tools, which, applied to a scikit-learn object, empowered dumping/loading of model and pipelines. The gold standard, named Pickle (or cPickle for the afficionados), already addressed serialization, in a different than frameworks like ONNX do nowadays. A question we could ask then : Is model serialization enough for modern machine learning projects ? Short and simple : no. Indeed, if serialization is THE issue the community started with, ONNX capabilities go far beyond that scope ..
But for now let’s define the main features, and limits, encountered when one would get the strange idea to dive into the wondrous world of manual pickling on a production environment. Here is a non-exhaustive list:
- First of all, pickle is a protocol, specific to Python, and nothing but Python. Models persisted to disk via Pickle can only get re-loaded in a program via Pickle, hence in Python. This is not a big issue if your application is all in Python but can become tedious if important changes occur.
- Latencies can be observed, especially when pickling/unpickling specific objects, see Matthew Rocklin’s post on this topic. In some cases Pickle lacks of information on the inner aspects of the object, and is left with a default non-optimal behaviour. This has no straight comparison with a typical ONNX workflow (see next paragraph), where serialization is not performed very frequently.
- On a cluster, no assumption is made on the locality of the object VS the data fed in. Object code can be shipped somewhere in the network while data relies far away from it. For this specific purpose was created cloudpickle
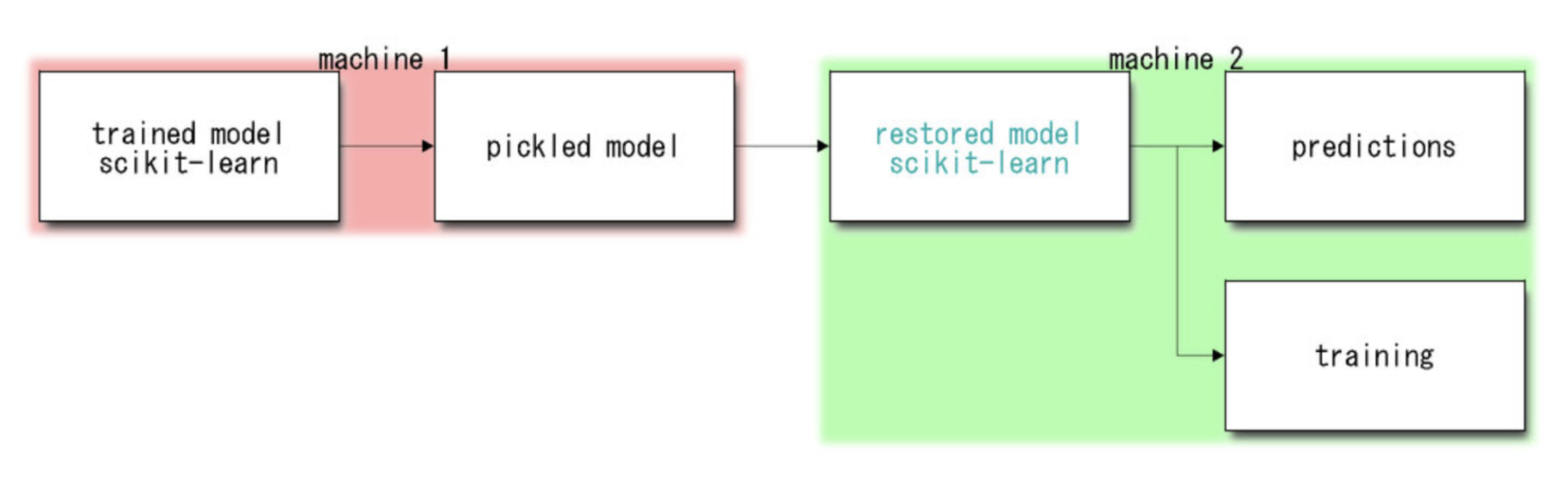
A common dump/load workflow with scitkit-learn and Pickle
On the other hand, Pickle allows us not only to dump our model, but also to reload it, entirely. This gives us the ability to re-train the model, and potentially re-dump it somewhere later.
sklearn – ready for batch predictions
scikit-learn, as we know, is really good at making predictions. Once the model fitted, it is easy to get a new piece of data and send it to the `predict` function. Unfortunatly, scikit-learn’s design make things not really optimal in some specific use-cases.
Indeed, the predict function is made to receive a numpy array, containing many elements, and return a numpy array, with the same number of elements : this is a batch prediction. This specific design gives you optimal runtimes when operating on a big number of entries : the more entries you have, the better you take advantage of vectorization.
On embedded systems for example, it is not always a good idea to buffer elements from a « stream“ before sending them to the predictmethod. This typical receive/buffer/predict workflow is not something to be considered in some cases. What could be done instead is sending every new entry into the mode/pipe, to get its corresponding predictions, in an online fashion.
sklearn-ONNX may be the solution for that.
sklearn-ONNX – online predictions and compatibility
From the necessity of having models from scikit-learn to run on different types of hard-wares (GPUs being an example), added to the growing need to provide fast on-line predictions, was born the sklearn-ONNX connector. The main role of sklearn-ONNX is basically to expose scikit-learn classes to the ONNX runtime, via object persistence in a .proto file (see the ONNX intermediate representation spec). Generally, models are persisted only once, with the single aim of being ported to a different environment.
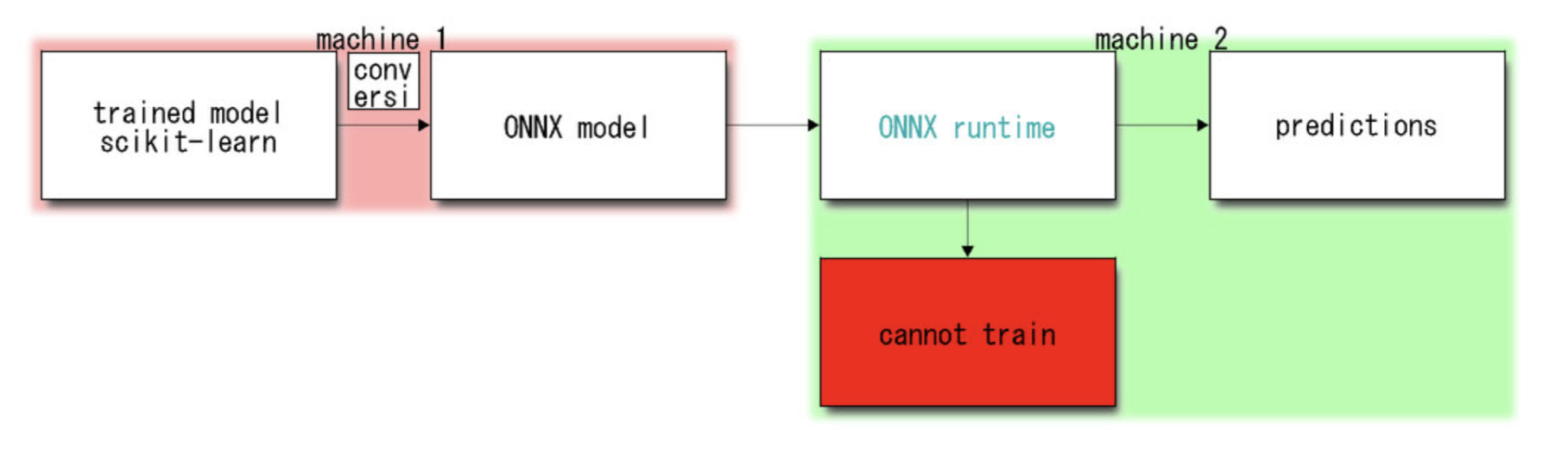
ONNX runtime, accessible thanks to the connector
sklearn-ONNX also gives us the opportunity to benchmark pure sklearn version VS skelarn-ONNX version when performing predictions one-by-one. Even if the benchmark is done in Python, this gives us a rough idea of what could be obtained in other environments. Here is a small example with a RandomForest Classifier.

Jumping to the conclusion :
- Pickle is great if you want to keep the ability re-train your model, or simply get an intermediate of your objects in a pure Python environment. It is currently used by popular librairies such as MLFLow in order to store/version models and pipelines
- sklearn-ONNX, on its side, is overwhelming when we need to use the same model on different physical environments, and generally performs extremely well at making predictions one by one. The main use case we keep in mind from sklearn-ONNX is porting a scikit-learn model on an embedded system, for low-consumption online predictions, potentially in C++.
Parallelism, memory, low-level optimizations – Jérémie de Boisberranger (INRIA)
Next to come is a presentation by Jérémie de Boisberranger, made up of common hints and guidelines to be followed when taking part in the development of high-performances tools like scikit-learn
Data types, still and always
A common underrated aspect when datascientists write code is datatypes, typically when dealing with high level APIs such as pandas, which provide type-inferrence.
We start though with a simple yet relevant example on a column of floating-point numbers represented either on 64 or 32 bits. Low levels instructions, at the level of SIMD register (Single Instruction Multiple Data) usually take advantage of data partitioning, by powers of 2. In the example below, the 32 bits version will offer better partitioning, simply by storing more data in a single register.
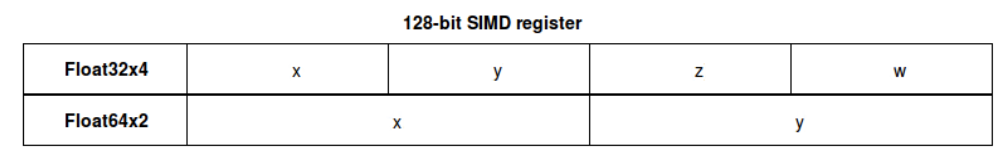
For 128 bits available on the register, we can store 4×32 bits, against 2×64 if we don’t provide the optimal data-type
Besides using 2x less RAM for the exact same number of entries in our DataFrame, the run time will also be impacted, because single vectorized operations will benefit from more data being shared (eg. matrix multiplication is a vanilla example)
Quick conclusion : We cannot rely on high level APIs to magically infer the optimal types for a unique use-case. Data scientists are responsible for their data, which includes providing the optimal types.
Intermediate results storage – just passing by
A growing number of methods in Machine Learning require storing intermediate values.
This holds true in unsupervised learning, for example in the K-Means algorithm, where it is necessary to store results at some points, like the pairwise-distances between each point in the data and the closest centroid. For this type of computation, multiple subparts of the data must be selected one by one to proceed.
Finding the optimal solution becomes finding an optimal size for this sub-part. If the value is too high, part of the data will end up being constantly exchanged between RAM and caches (L1, L2, L3). It the value is too low, everything will holds in the cache (faster computation) but a bigger number of iterations will be required. The RAM consumption will decrease, but the running time increase …
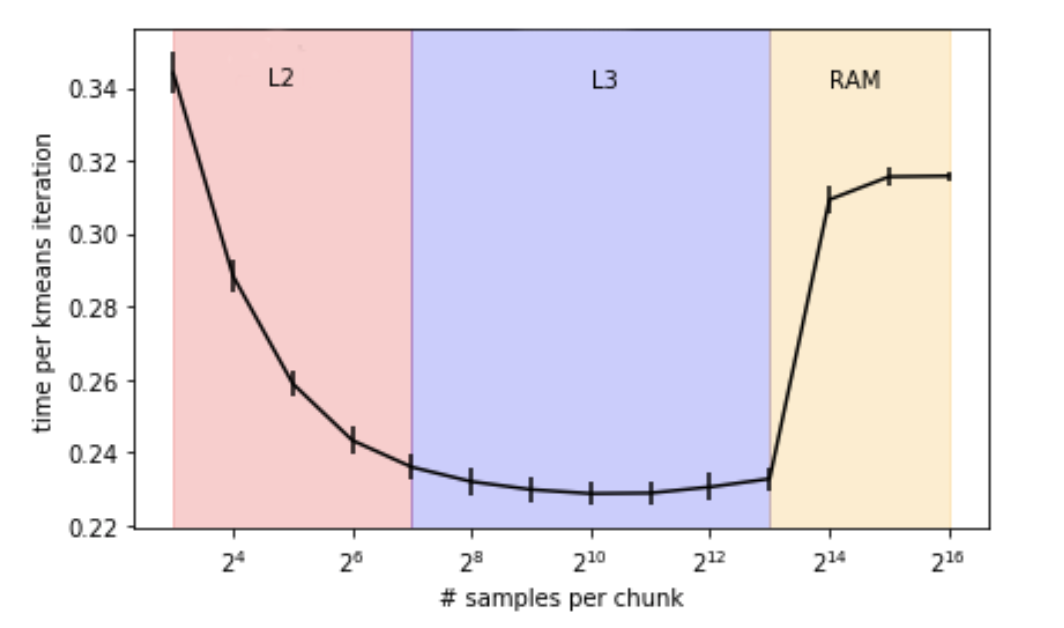
Note that an alternate version of K-Means, named mini-batch K-means, avoid this type of problems, partly at least, by running on random samples of the original data, which reduces the number of pair-wise distances to be computed, and therefore the size of the candidate intermediate results.
Over-subscription
In scikit-learn, most of the operations that can easily be parallelised in a naive fashion (embarassingly parallel for loops), are managed via joblib, a high-level API created and maintained by Olivier Grisel.
Joblib objects manage their own resources, and therefore the number of threads and processes, allocated via the n_jobs parameter. Nonetheless, Joblib is a high level tool, mainly in charge of executing operations which are for the most part implemented by the user himself; a function to be run on a sample of data for example.
Lower-levels APIs (NumPy, Scipy, …), on their side, also internally manage their own resources, and more specifically their interactions with the BLAS, via different libraries : OpenBLAS, MKL, … This inner parallelism, far away from what Joblib can acknowledge, is the problem …
A short example
An example provided in the notebook, gives an idea of what happens in a pretty standard use-case, producing side-effects that can lead to disastrous drops in the runtime. At instantiation time, the user asks the scikit-learn class to use 4 jobs, simply by setting n_jobs to 4. If the type of scheduler is left to default, Joblib will throw n_jobs processes to go through the computation. Somewhere else in the codebase of the class, calls to the NumPy API are made, whci themselves trigger calls to the BLAS, which will try to execute its own instructions in parallel, managing its own number of threads, independently from the n_jobs parameter chosen by the user …
If we run this computation with n_jobs equal to the number of available CPUs, and do not tell the BLAS to limit itself in the way it allocates ressources, we end end up trying to benefit from an architecture we do not physically possess : this is a case of over-subscription. Hopefully for us, the number of ressources accessible by the BLAS can be controlled, via a dedicated environment variable. For the MKL BLAS for example, MKL_NUM_THREADS=1 will limit the number of threads to 1, and let Joblib properly access all the available ressources.
Finally, recent version of Joblib natively support this kind of cases, via threadpoolctl. Users of the latest versions can use Joblib objects without paying much attention of over-subscription.
Distributed GPU Machine Learning with RAPIDS and Dask – Peter Entschev (Nvidia)
Dask : Long story short
Dask started at Continuum Analytics a few years ago. Back in 2015, Anaconda Inc was looking for a way to get complex algorithms implemented in python all set up on multicore architectures, or potentially on clusters of machines. The solution had to be generic enough to ingest any kind of Python code, while providing transparent layers on top of the tools that made Python such a great ecosystem for scientific computations.
The building block of Dask is the delayed decorator, which can set any python function as a node in a DAG, and makes it a candidate for later computation, by a scheduler.
For the past few year more and more blog posts emerged, not only from its creators (Matthew Rocklin and co) but from a fastly growing community of users willing to take advanced DataScience algorithm to big clusters. Its reliability, its well furnished documentation and its strong compatiilty with the existing blocks (pandas, NumPy, …) makes it the gold standard in terms of parallel computing in the Python world.
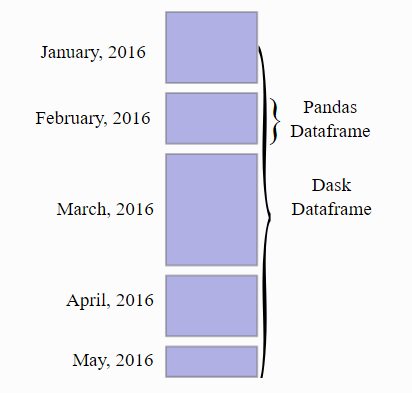
The most commonly used data structure in Dask is probably the dask.DataFrame, which can be seen as a smart way to trigger computations on many pandas.DataFrames.
In late 2017, the first threads appeared on github that mentionned the possibility of dropin replacement for pandas.DataFrame by DataFrames of a new kind, backed by GPUs routines.
In 2018, the maintainer of Dask, Matthew Rocklin, left Anaconda to join Nvidia …
RAPIDS
RAPIDS is a simple project, with a simple goal : recreated the existing (CPU) scientific computing APIs, on GPUs, to be able to run the same computation on Nvidia GPUs chips.
RAPIDS reasons in a bottom-up approach : if we can re-implement all the building blocks leading to smart algorithms (like we have in scikit-learn), then we can, going up the stack, re-implement the most famous algorithms with only minimal change in the code base.
RAPIDS is decoupled into many sub-projets, all referring to different « levels » in the stack. From low to high level, we have
- CUDA : a C library for programming on GPU chips
- cuPy – a NumPy like API, built on top of CUDA
- cuDF– a pandas like API, for data preparation. Only a subset of the pandas API is currently supported.
- cuML – a scikit-learn like API : for accelerated machine learning
- dask-cuDF – to make dask.DataFrame internally manage cuDf.DataFrames, and run many GPUs, either on a single machine or on a cluster of machines. Because they rely on cuDf, they only support a subset of the dask.DataFrame API
Low level considerations : The curse of modern data science
Even after having spend a few years working for a consulting company like Quantmetry, it is not that easy to identify what makes consultants productive on a daily basis, knowing they will have to face different situations on different missions.
For the past few years DataScience was THE brand new thing every company wanted to step in, most of the time resulting in launching Proof Of Concepts (POC). These projects are usually short (a few months), and shall demonstrate value very quickly.
Data Scientists don’t care about low level optimizations
This project-level considerations results in Data Scientists to seek for langages and technologies that leverage fast prototyping and experiment over the existing data, without knowing much about the software (compilation, memory management, testing … ). This way of delivering projects is conceptually the opposite way software products are developed in the classical computer-science world, where programming itself is considered as an art, and langages are carefully chosen for their specificities.
Reduced delivery deadlines, added to the increasing availability of a variety of compute units on the cloud (including GPUs), make RAPIDS a good candidate to modern Data Science applications.
But how does RAPIDS operate ?
Different hardwares, common interfaces
Again, the whole point of interfacing these libraries together is to be able to switch between CPU and GPU with low impact on the code.
On the GPU side, the CUDA array interface allows sharing GPU arrays between libraries, starting from CuPy and getting to broader applications like PyTorch.
On the CPU side, the Numpy Array Function allows interoperability between NumPy-like libraries, CuPy being one of them.
Passing a CuPy array to a structure requiring Numpy-like input will only require 2 lines of code to be changed
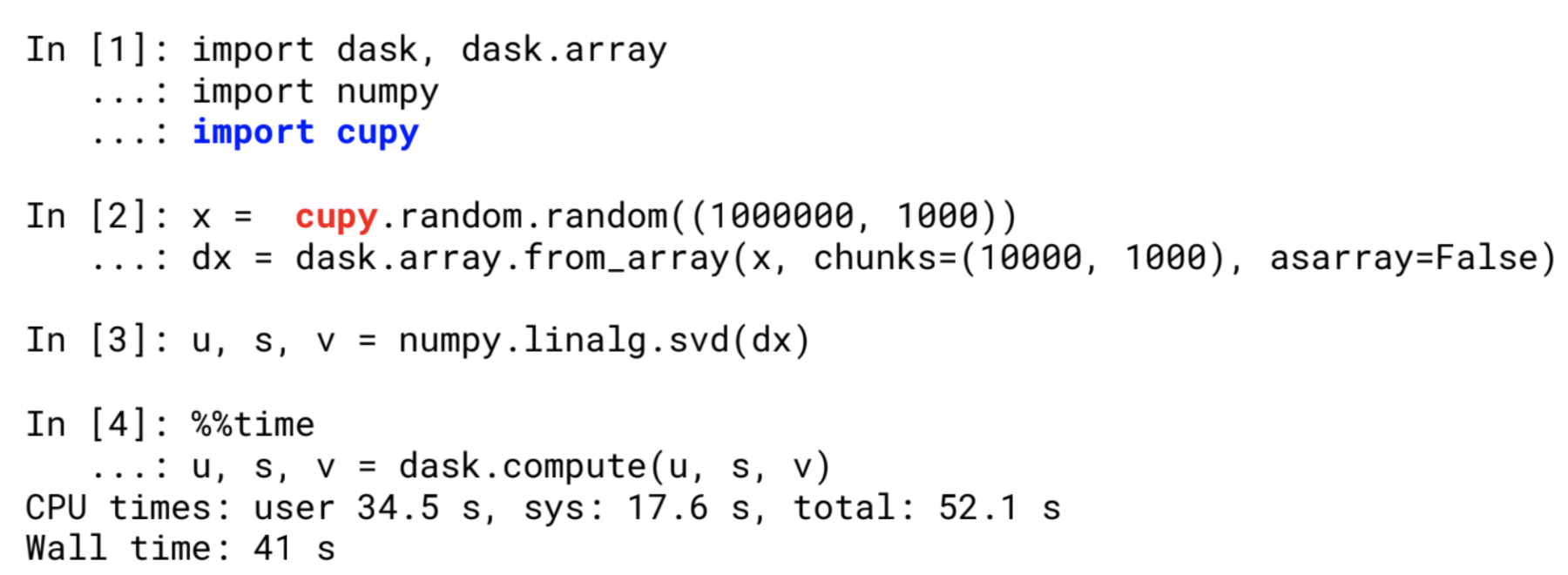
Dask.array respects the Numpy Array Function interface, and can take a cupy array as input
CuML Roadmap
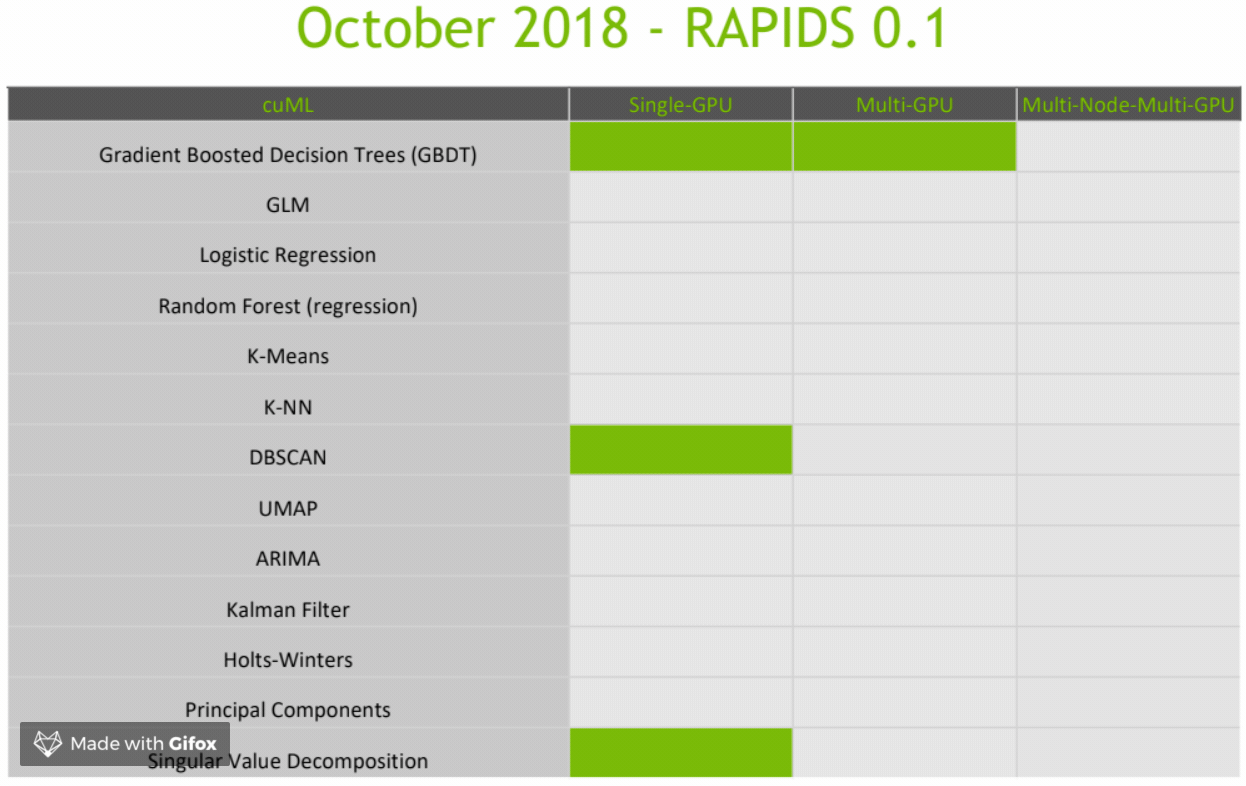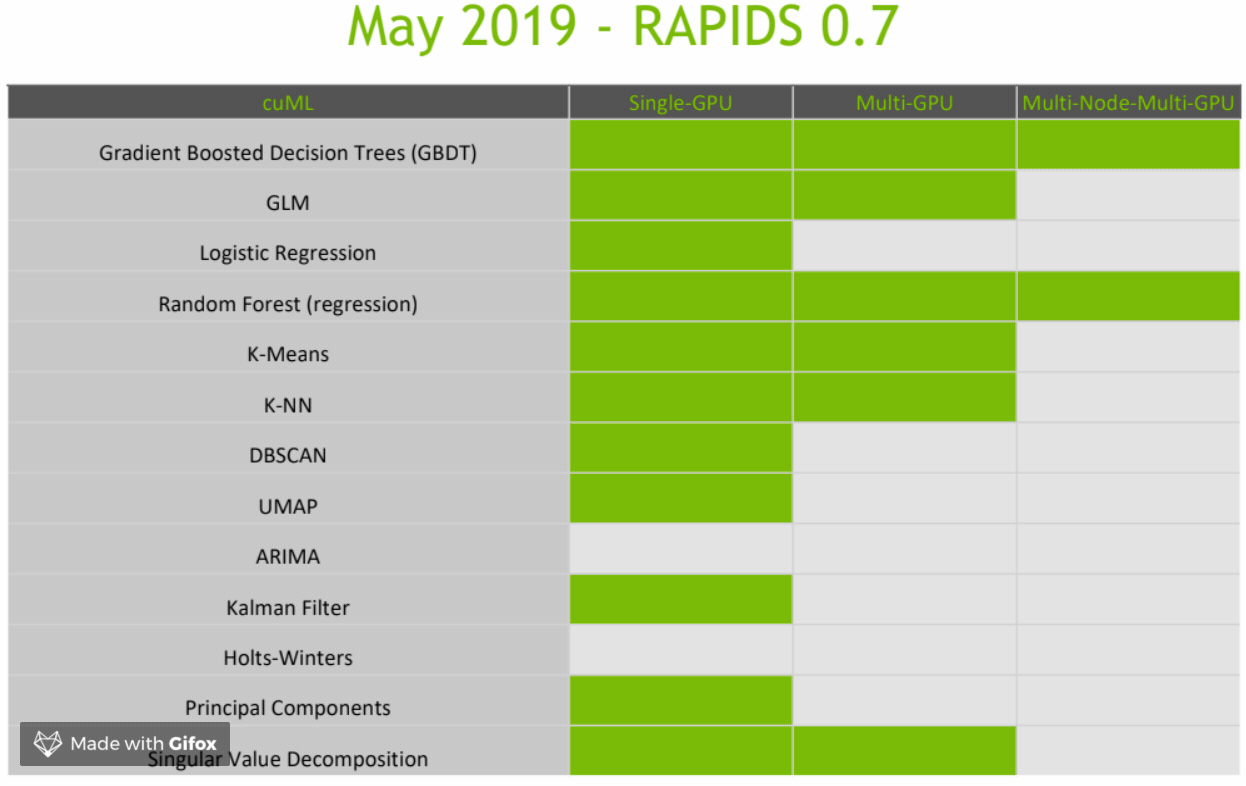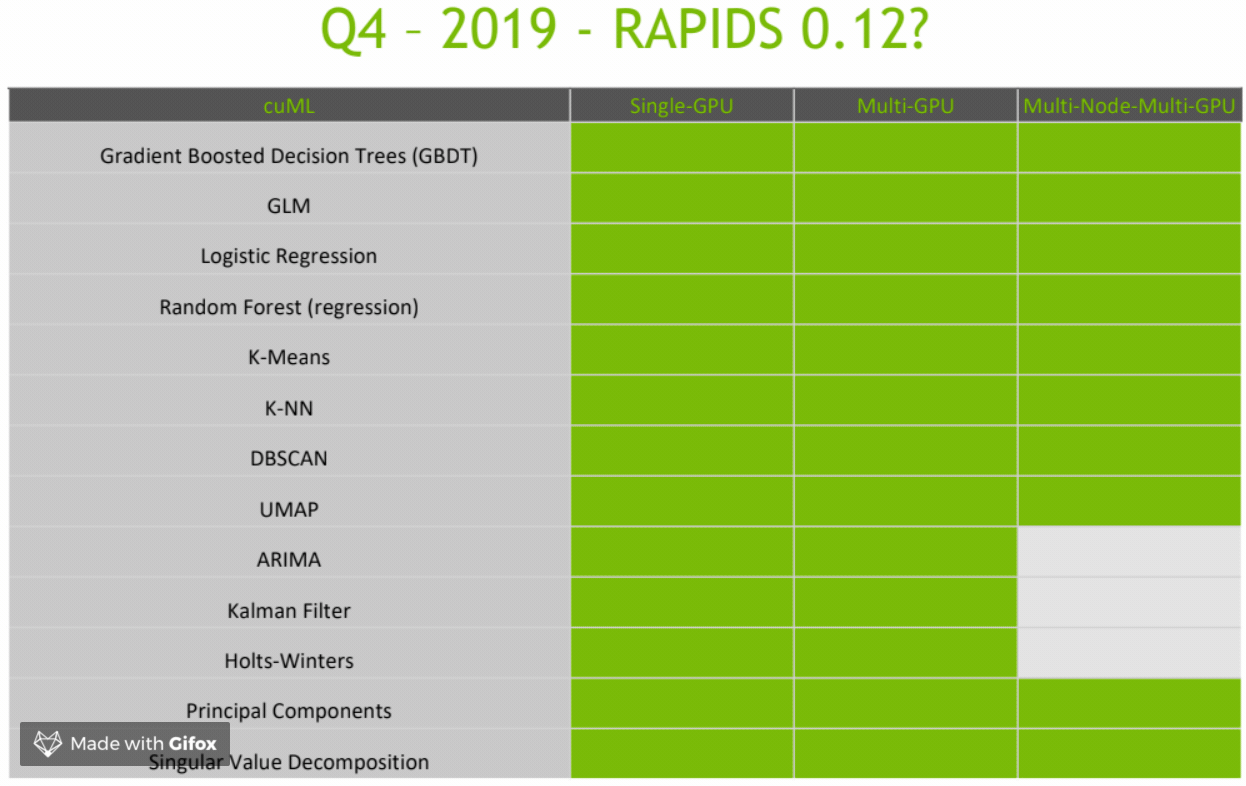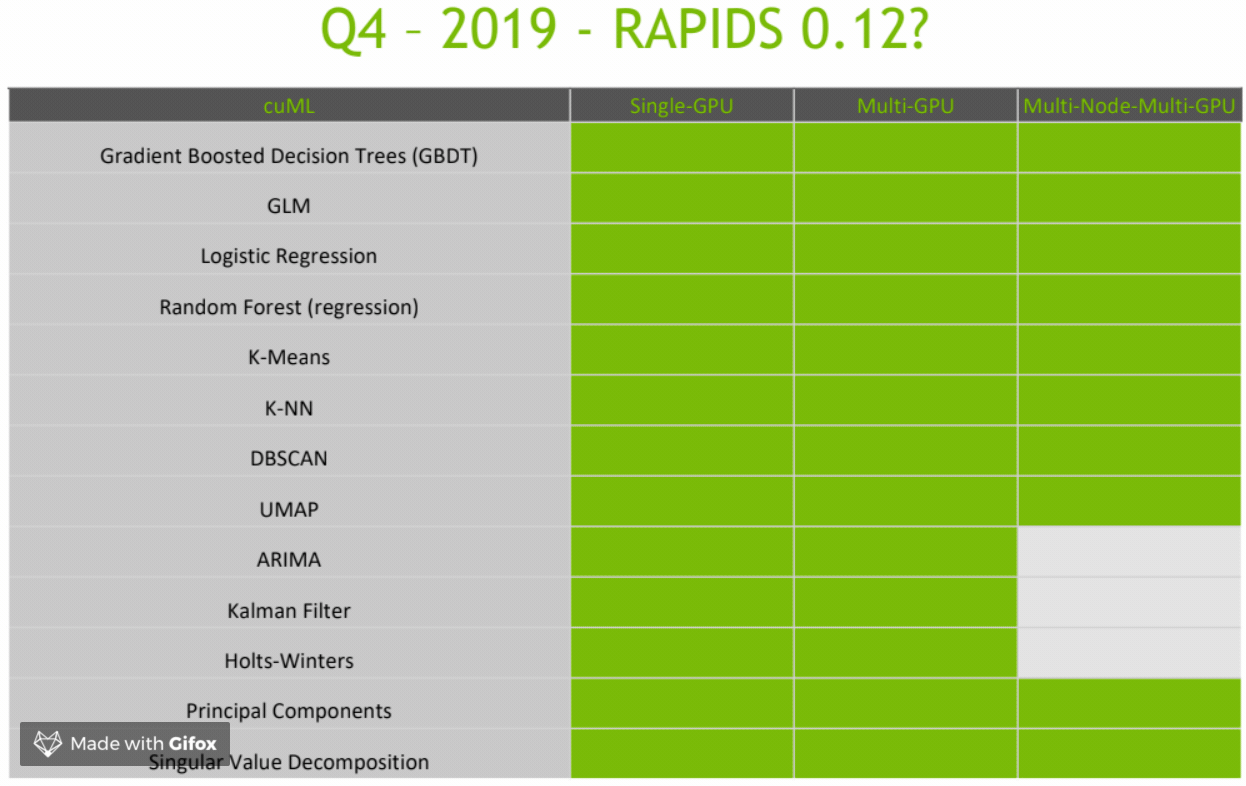
Final Thoughts
